Part 3: Tag Maps Clustering and Topic Heat Maps
Workshop: Social Media, Data Analysis, & Cartograpy, WS 2022/23
Alexander Dunkel, Madalina Gugulica, Institute of Cartography, TU Dresden
This is the third notebook in a series of four notebooks:
- Introduction to Social Media data, jupyter and python spatial visualizations
- Introduction to privacy issues with Social Media data and possible solutions for cartographers
- Specific visualization techniques example: TagMaps clustering
- Specific data analysis: Topic Classification
Open these notebooks through the file explorer on the left side.
- Please make sure that "03_tagmaps_env" is shown on the top-right corner. If not, click & select.
Introduction: Privacy & Social Media¶
The task in this notebook is to extract and visualize common areas or regions of interest for a given topic from Location Based Social Media data (LBSM).
On Social Media, people react to many things, and a significant share of these reactions can be seen as forms of valuation or attribution of meaning to to the physical environment.
Challenges
However, for visualizing such values for whole cities, there are several challenges involved:
- Identify reactions that are related to a given topic:
- Topics can be considered at different levels of granularity
- and equal terms may convey different meanings in different contexts.
- Social Media data is distributed very irregularly.
- Data peaks in urban areas and highly frequented tourist hotspots,
- while some less frequented areas do not feature any data at all.
- When visualizing spatial reactions to topics, this unequal distribution must be taking into account, otherwise maps would always be biased towards areas with higher density of information.
- For this reason, normalizing results is crucial.
- The usual approach for normalization in such cases is that local distribution is evaluated against the global (e.g. spatial) distribution data.
Tag Maps Package
- The Tag Maps package was developed to cluster tags according to their spatial area of use
- Currently, the final visualization step of Tag Maps clustering (placing labels etc.) is only implemented in ArcMap
- Herein, we instead explore cluster results using Heat Maps and other graphics in python
- In the second part of the workshop, it will be shown how to visualize the actual Tag Maps
Where is my custom area/tag map?
- Below, you can chose between different regions/areas to explore in this notebook, provided based on student input
- The final Tag Map visualization step will be done in the second part of the workshop
- This will be done in ArcMap. However, it is possible to visualize TagMaps in jupyter Lab, too. The notebook is not yet ready - have a look at the work in progress here
Important:
- The privacy-aware data structure shown in the second Notbook has not been implemented yet in tag maps
- We will be working with RAW data instead
- Please:
- Do not share the original data
- Remove original data after the workshop
Preparations¶
Define output directory
from pathlib import Path
OUTPUT = Path.cwd() / "out"
Temporary fix to prevent proj-path warning:
import sys, os
os.environ["PROJ_LIB"] = str(Path(sys.executable).parents[1] / 'share' / 'proj')
Create 01_Input folder, which will be used for loading data.
INPUT = Path.cwd() / "01_Input"
Load the TagMaps package, which will serve as a base for filtering, cleaning and processing noisy social media data
from tagmaps import TagMaps, EMOJI, TAGS, TOPICS, LoadData, BaseConfig
from tagmaps.classes.shared_structure import ItemCounter
%load_ext autoreload
%autoreload 2
import sys
module_path = str(Path.cwd().parents[0] / "py")
if module_path not in sys.path:
sys.path.append(module_path)
from modules import preparations
preparations.init_packages()
from modules import tools
# enable ignore shapely deprecation warnings
import warnings
from shapely.errors import ShapelyDeprecationWarning
warnings.filterwarnings("ignore", category=ShapelyDeprecationWarning)
Load Data & Plot Overview¶
Retrieve sample LBSN CSV data:
source = "GrosserGarten.zip"
# source = "Istanbul.zip"
# source = "Lindau.zip"
# source = "tumunich.zip"
# source = "ArgentinaCarhue.zip"
# source = "MachuPicchu.zip"
# source = "Rudolf-Harbig-Stadion.zip"
Select Input
- Based on requests, we have provided several sources of data
- Choose one of the input source above
- The default input data is for the Großer Garten in Dresden
%%time
# clean any data first
tools.clean_folders([INPUT])
sample_url = tools.get_sample_url()
lbsn_dd_csv_uri = f'{sample_url}/download?path=%2F&files='
tools.get_zip_extract(
uri=lbsn_dd_csv_uri,
filename=source,
output_path=INPUT,
write_intermediate=True)
Initialize tag maps from BaseConfig
tm_cfg = BaseConfig()
Optionally, filter data per origin or adjust the number of top terms to extract:
tm_cfg.filter_origin = None
tm_cfg.max_items = 3000
tm_opts = {
'tag_cluster':tm_cfg.cluster_tags,
'emoji_cluster':tm_cfg.cluster_emoji,
'location_cluster':tm_cfg.cluster_locations,
'output_folder':tm_cfg.output_folder,
'remove_long_tail':tm_cfg.remove_long_tail,
'limit_bottom_user_count':tm_cfg.limit_bottom_user_count,
'max_items':tm_cfg.max_items,
'topic_cluster':True}
tm = TagMaps(**tm_opts)
**kwargs syntax?
- Some functions take a long list of (keyword) arguments
- These keyword arguments can be passed to the function using a dictionary
- The
**syntax tells python to unpack keyword arguments (kwargs) from a dictionary
a) Read from original data¶
Read input records from csv
from IPython.display import clear_output
%%time
input_data = LoadData(tm_cfg)
with input_data as records:
for ix, record in enumerate(records):
tm.add_record(record)
if (ix % 1000) != 0:
continue
# report every 1000 records
clear_output(wait=True)
print(f'Loaded {ix} records')
input_data.input_stats_report()
tm.prepare_data()
b) Optional: Write (& Read) cleaned output to file¶
Output cleaned data to Output/Output_cleaned.csv,
clean terms (post_body and tags) based on the top (e.g.) 1000 hashtags found in dataset.
tm.lbsn_data.write_cleaned_data(panon=True)
Have a look at the output file.
import pandas as pd
file_path = Path.cwd() / "02_Output" / "Output_cleaned.csv"
display(pd.read_csv(file_path, nrows=5).head())
print(f'{file_path.stat().st_size / (1024*1024):.02f} MB')
Data Minimization
- The cleaned output file is significantly smaller than original data
- It includes only a small subset of original terms, based on filtering of frequently used terms
- As a minor precaution, IDs, such as guid or user_guid, have been pseudonymized.
- In reality, such pseudonymization will be easy to reverse, and thus only provides a minor protection to misuse of data
Read from pre-filtered data
Read data form (already prepared and filtered) cleaned output
%%time
tm.prepare_data(
input_path=file_path)
tm.global_stats_report()
tm.item_stats_report()
Topic selection & Tag Clustering¶
The next goal is to select reactions to given topics. TagMaps allows selecting posts for 4 different types:
- TAGS (i.e. single terms)
- EMOJI (i.e. single emoji)
- LOCATIONS (i.e. named POIs or coordinate pairs)
- TOPICS (i.e. list of terms)
Set basic plot to notebook mode and disable interactive plots for now
import matplotlib.pyplot as plt
import matplotlib as mpl
%matplotlib inline
mpl.rcParams['savefig.dpi'] = 120
mpl.rcParams['figure.dpi'] = 120
We can retrieve preview maps for the TOPIC dimension
by supplying a list of terms.
For example "park", "green" and "grass" should
give us an overview where such terms are used
in our sample area.
Custom topic
- Try to process first with the supplied set of terms
- Afterwards, choose your own set of terms to affect visualizations below
- You can repeat the process for different topics by starting in this cell again
nature_terms = ["park", "grass", "nature"]
fig = tm.get_selection_map(
TOPICS, nature_terms)
urban_terms = ["strasse", "city", "shopping"]
fig = tm.get_selection_map(
TOPICS, urban_terms)
HDBSCAN Cluster¶
We can visualize clusters for the selected topic using HDBSCAN.
The important parameter for HDBSCAN is the cluster distance,
which is chosen automatically by Tag Maps given the current scale/extent of analysis.
# tm.clusterer[TOPICS].cluster_distance = 150
fig = tm.get_cluster_map(
TOPICS, nature_terms)
Manually select cluster distance
- TagMaps automatically selects a suitable cluster distance
- For the Großer Garten dataset, this distance is 203 meters
- We can select a smaller cluster distance, to get finer grained picture of topic clusters.
- In the cell above, uncomment the first line and re-run the cell
- Try other cluster distances to see how the output changes.
We can get a map of clusters and cluster shapes (convex and concave hulls).
fig = tm.get_cluster_shapes_map(
TOPICS, nature_terms)
Behind the scenes, Tag Maps utilizes the Single Linkage Tree from HDBSCAN to cut clusters at a specified distance. This tree shows the hierarchical structure for our topic and its spatial properties in the given area.
fig = tm.get_singlelinkagetree_preview(
TOPICS, nature_terms)
Cluster centroids¶
Similarly, we can retrieve centroids of clusters. This shows again the unequal distribution of data:
fig = tm.clusterer[TOPICS].get_cluster_centroid_preview(
nature_terms, single_clusters=True)
Heat Maps¶
- Visualization of clustered tagmaps data is possible in several ways.
- In the second workshop (end of January), we are using ArcMap, to create labeled maps from clustered data
- The last part in this notebook will be to use Kernel Densitry Estimation (KDE) to create a Heat Map for selected topics.
Further information: KDE
- Some of the code below was adapted from plot species example
- A more thorough tutorial is provided here
- The Python Data Science Handbook also contains a section on KDE
Load additional dependencies
import numpy as np
from sklearn.neighbors import KernelDensity
Load Flickr data only¶
Instagram, Facebook and Twitter are based on place-accuracy, which is unsuitable for the Heat Map graphic.
We'll work with Flickr data only for the Heat Map.
- 1 = Instagram
- 2 = Flickr
- 3 = Twitter
Reload data, filtering only Flickr
tm_cfg.filter_origin = "2"
tm = TagMaps(**tm_opts)
%%time
input_data = LoadData(tm_cfg)
with input_data as records:
for ix, record in enumerate(records):
tm.add_record(record)
if (ix % 1000) != 0:
continue
# report every 1000 records
clear_output(wait=True)
print(f'Loaded {ix} records')
input_data.input_stats_report()
Get Topic Coordinates¶
The distribution of these coordinates is what we want to visualize.
tm.prepare_data()
tm.init_cluster()
Topic selection
topic = "grass-nature-park"
points = tm.clusterer[TOPICS].get_np_points(
item=topic,
silent=True)
Why a different syntax? (term-term-term)
- The API of
tagmapspackage is still in an early stage - The above function uses the internal representation of topics
Get All Points¶
all_points = tm.clusterer[TOPICS].get_np_points()
For normalizing our final KDE raster, we'll process both, topic selection points and global data distribution (e.g. all points in the dataset).
points_list = [points, all_points]
- The input data is a simply list (as a
numpy.ndarray) of decimal degree coordinates - each entry represents a single user that published one or more posts at a specific coordinate
print(points[:5])
print(f'Total coordinates: {len(points)}')
Data projection¶
- For faster KDE, we project data from WGS1984 (
epsg:4326) to UTM - this allows us to directly calculate in euclidian space.
- The TagMaps package automatically detected the most suitable UTM coordinate system,
- for the Großer Garten sample data, this is Zone 33N (
epsg:32633)
Project lat/lng to UTM 33N (Dresden) using Transformer:
%%time
for idx, points in enumerate(points_list):
points_list[idx] = np.array(
[tm.clusterer[TOPICS].proj_transformer.transform(
point[0], point[1]
) for point in points])
crs_wgs = tm.clusterer[TOPICS].crs_wgs
crs_proj = tm.clusterer[TOPICS].crs_proj
print(f'From {crs_wgs} \nto {crs_proj}')
print(points_list[0][:5])
Calculating the Kernel Density¶
To summarize sklearn, a KDE is executed in two steps, training and testing:
Machine learning is about learning some properties of a data set and then testing those properties against another data set. A common practice in machine learning is to evaluate an algorithm by splitting a data set into two. We call one of those sets the training set, on which we learn some properties; we call the other set the testing set, on which we test the learned properties.
The only attributes we care about in training are lat and long.
- Stack locations using np.vstack, extract specific columns with [:,column_id]
- reverse order: lat, lng
- Transpose to Rows (.T), easier in python ref
xy_list = list()
for points in points_list:
y = points[:, 1]
x = points[:, 0]
xy_list.append([x, y])
xy_train_list = list()
for x, y in xy_list:
xy_train = np.vstack([y, x]).T
xy_train_list.append(xy_train)
print(xy_train)
Get bounds from total data
Access min/max decimal degree bounds object of clusterer and project to UTM33N
lim_lng_max = tm.clusterer[TAGS].bounds.lim_lng_max
lim_lng_min = tm.clusterer[TAGS].bounds.lim_lng_min
lim_lat_max = tm.clusterer[TAGS].bounds.lim_lat_max
lim_lat_min = tm.clusterer[TAGS].bounds.lim_lat_min
# project WDS1984 to UTM
topright = tm.clusterer[TOPICS].proj_transformer.transform(
lim_lng_max, lim_lat_max)
bottomleft = tm.clusterer[TOPICS].proj_transformer.transform(
lim_lng_min, lim_lat_min)
# get separate min/max for x/y
right_bound = topright[0]
left_bound = bottomleft[0]
top_bound = topright[1]
bottom_bound = bottomleft[1]
Create Sample Mesh
Create a grid of points at which to predict.
xbins=100j
ybins=100j
xx, yy = np.mgrid[
left_bound:right_bound:xbins,
bottom_bound:top_bound:ybins]
xy_sample = np.vstack([yy.ravel(), xx.ravel()]).T
Generate Training data for Kernel Density.
- The largest effect on the final result comes from the chosen bandwidth for KDE - smaller bandwidth mean higher resultion,
- but may not be suitable for the given density of data (e.g. results with low validity).
- Higher bandwidth will produce a smoother raster result, which may be too inaccurate for interpretation.
Modify bandwidth parameter
- The bandwidth affects the distance at which points are considered during KDE
- Test all cells below first with the given bandwidth (200 meters)
- Afterwards, come back to this cell and adjust backwidth
- Compare the effect on visualization
kde_list = list()
for xy_train in xy_train_list:
kde = KernelDensity(
kernel='gaussian', bandwidth=200, algorithm='ball_tree')
kde.fit(xy_train)
kde_list.append(kde)
score_samples() returns the log-likelihood of the samples
%%time
z_list = list()
for kde in kde_list:
z_scores = kde.score_samples(xy_sample)
z = np.exp(z_scores)
z_list.append(z)
Remove values below zero,
these are locations where selected topic is underrepresented, given the global KDE mean
for ix in range(0, 2):
z_list[ix] = z_list[ix].clip(min=0)
Normalize z-scores to 1 to 1000 range for comparison
for idx, z in enumerate(z_list):
z_list[idx] = np.interp(
z, (z.min(), z.max()), (1, 1000))
Subtact global zscores from local z-scores (our topic selection)
norm_val:
- weight of global z-score
- lower means less normalization effect, higher means stronger normalization effect
- range:
0to1
norm_val = 0.5
z_orig = z_list[0]
z_is = z_list[0] - (z_list[1]*norm_val)
z_results = [z_orig, z_is]
Modify norm_val parameter
- The normalization affects the the strength at which global values will have an effect on local values (the topic)
- Test all cells below first with the given normalization (0.5)
- Afterwards, come back to this cell and adjust normalization
- Compare the effect on visualization
Reshape results to fit grid mesh extent
for idx, z_result in enumerate(z_results):
z_results[idx] = z_results[idx].reshape(xx.shape)
Plot original and normalized meshes
from matplotlib.ticker import FuncFormatter
def y_formatter(y_value, __):
"""Format UTM to decimal degrees y-labels for improved legibility"""
xy_value = tm.clusterer[
TOPICS].proj_transformer_back.transform(
left_bound, y_value)
return f'{xy_value[1]:3.2f}'
def x_formatter(x_value, __):
"""Format UTM to decimal degrees x-labels for improved legibility"""
xy_value = tm.clusterer[
TOPICS].proj_transformer_back.transform(
x_value, bottom_bound)
return f'{xy_value[0]:3.1f}'
# a figure with a 1x2 grid of Axes
fig, ax_lst = plt.subplots(1, 2, figsize=(10, 3))
for idx, zz in enumerate(z_results):
axis = ax_lst[idx]
# Change the fontsize of minor ticks label
axis.tick_params(axis='both', which='major', labelsize=7)
axis.tick_params(axis='both', which='minor', labelsize=7)
# set plotting bounds
axis.set_xlim(
[left_bound, right_bound])
axis.set_ylim(
[bottom_bound, top_bound])
# plot contours of the density
levels = np.linspace(zz.min(), zz.max(), 15)
# Create Contours,
# save to CF-variable so we can later reference
# it in colorbar hook
CF = axis.contourf(
xx, yy, zz, levels=levels, cmap='viridis')
# titles for plots
if idx == 0:
title = f'Original KDE for topic {topic}\n(Flickr Only)'
else:
title = f'Normalized KDE for topic {topic}\n(Flickr Only)'
axis.set_title(title, fontsize=11)
axis.set_xlabel('lng', fontsize=10)
axis.set_ylabel('lat', fontsize=10)
# plot points on top
axis.scatter(
xy_list[1][0], xy_list[1][1], s=1, facecolor='white', alpha=0.05)
axis.scatter(
xy_list[0][0], xy_list[0][1], s=1, facecolor='white', alpha=0.1)
# replace x, y coords with decimal degrees text (instead of UTM dist)
axis.yaxis.set_major_formatter(
FuncFormatter(y_formatter))
axis.xaxis.set_major_formatter(
FuncFormatter(x_formatter))
if idx > 0:
break
# Make a colorbar for the ContourSet returned by the contourf call.
cbar = fig.colorbar(CF,fraction=0.046, pad=0.04)
cbar.set_title = ""
cbar.sup_title = ""
cbar.ax.set_ylabel(
"Number of \n User Post Locations (UPL)", fontsize=11)
cbar.ax.set_title('')
cbar.ax.tick_params(axis='both', labelsize=7)
cbar.ax.yaxis.set_tick_params(width=0.5, length=4)
Depending on the data source and chosen topic, the two maps above will differ more or less.
For the Großer Garten Example and and nature topic,
the normalized map features little difference to the original KDE,
likely because there are little false positives in the surrounding area for nature reactions.
Combine HDBSCAN and KDE¶
For the final map, we want to combine the different data we collected.
This means adding cluster shapes from HDBSCAN and generating
a standalone interactive and external html for sharing.
def get_poly_patch(polygon):
"""Returns a matplotlib polygon-patch from shapely polygon"""
patch = PolygonPatch(
polygon, fc='#000000',
ec='#999999', fill=False,
zorder=1, alpha=0.7)
return patch
def add_patch(polygon, axis):
xs, ys = polygon.exterior.xy
axis.fill(xs, ys, alpha=0.5, fc='none', ec='white')
Experimental code below
- Frequently, you will mix more production-ready code with experimental code cells in jupyter
- The code cell below is an example of experimental code:
- Things may change often here
- Only few code comments are provided
- A good recommendation is to put more effort into code that you will frequently re-use
- Once methods reach a certain maturity, store those in separate modules (e.g. the
tools.pyused herein) - This allows you to re-use methods in other notebooks as well.
Modify cluster_distance parameter
- If you're working with other area datasets, adjust
cluster_distancebelow to a suitable distance (meters)
fig, axis = plt.subplots(1)
# Change the fontsize of minor ticks label
axis.tick_params(
axis='both', which='major', labelsize=7)
axis.tick_params(
axis='both', which='minor', labelsize=7)
# set plotting bounds
axis.set_xlim(
[left_bound, right_bound])
axis.set_ylim(
[bottom_bound, top_bound])
# plot contours of the density
levels = np.linspace(zz.min(), zz.max(), 25)
CF = axis.contourf(
xx, yy, zz, levels=levels, cmap='viridis')
title = (f'Normalized KDE for topic {topic} '
f'\n with HDBSCAN Cluster Shapes added '
f'\n(Flickr Only)')
axis.set_title(title)
axis.set_xlabel('lng', fontsize=10)
axis.set_ylabel('lat', fontsize=10)
# plot points on top
axis.scatter(
xy_list[0][0], xy_list[0][1],
s=1, facecolor='white', alpha=0.1)
axis.yaxis.set_major_formatter(
FuncFormatter(y_formatter))
axis.xaxis.set_major_formatter(
FuncFormatter(x_formatter))
# Make a colorbar for the ContourSet
# returned by the contourf call.
cbar = fig.colorbar(
CF,
label='Number of \n User Post Locations (UPL)',
fraction=0.046, pad=0.04)
cbar.set_title = ""
cbar.sup_title = ""
cbar.ax.set_title('')
cbar.ax.tick_params(axis='both', labelsize=7)
cbar.ax.yaxis.set_tick_params(width=0.5, length=4)
# Get Cluster Shapes from TagMaps
tm.clusterer[TOPICS].cluster_distance = 200
result = tm.clusterer[TOPICS].cluster_item(
item=topic,
preview_mode=True)
clusters = result.clusters
selected_post_guids = result.guids
cluster_guids = tm.clusterer[TOPICS]._get_cluster_guids(
clusters, selected_post_guids)
shapes = tm.clusterer[TOPICS]._get_item_clustershapes(
ItemCounter(topic, 0), cluster_guids.clustered)
for alpha_shape in shapes.alphashape:
add_patch(alpha_shape.shape, axis)
Overlay Raster in Geoviews for interactive display¶
import holoviews as hv
import geoviews as gv
import geoviews.feature as gf
from cartopy.crs import epsg
import geoviews.tile_sources as gts
hv.notebook_extension('bokeh')
Get cartopy projection from epsg string.
cc_proj = epsg(
crs_proj.split(":")[1])
Create contours from kde data
contours = gv.FilledContours(
(xx.T, yy.T, zz.T),
vdims='Number of \n User Post Locations (UPL)',
crs=cc_proj)
Create additional layer for HDBSCAN cluster shapes
shape_list = [shape.shape for shape in shapes.alphashape]
gv_shapes = gv.Polygons(
shape_list,
label=f'HDBSCAN city-wide Cluster Shapes for topic',
crs=cc_proj)
Prepare plotting
def set_active_tool(plot, element):
"""Enable wheel_zoom by default"""
plot.state.toolbar.active_scroll = plot.state.tools[0]
Initial zoom extent
- The Overlay will automatically extent to data dimensions, the polygons of HDBSCAN cover the city area
- Since we want the initial zoom to be zoomed in to the Großer Garten we need to supply xlim and ylim.
- The Bokeh renderer only support Web Mercator Projection
- we therefore need to project Großer Garten Bounds first from WGS1984 to Web Mercator.
- We'll also zoom in from original max extents by about 50%
Prepare projection from WGS1984 to Web Mercator
from pyproj import Transformer
crs_proj = f"epsg:4326"
crs_wgs = "epsg:3857"
# define Transformer ahead of time
# with xy-order of coordinates
proj_transformer = Transformer.from_crs(
crs_wgs, crs_proj, always_xy=True)
# also define reverse projection
proj_transformer_back = Transformer.from_crs(
crs_proj, crs_wgs, always_xy=True)
Zoom in to 50% of original data extent
xcrop = (lim_lng_max - lim_lng_min) / 4
ycrop = (lim_lat_max - lim_lat_min) / 4
lim_lng_min_crop = lim_lng_min + xcrop
lim_lng_max_crop = lim_lng_max - xcrop
lim_lat_min_crop = lim_lat_min + ycrop
lim_lat_max_crop = lim_lat_max - ycrop
Apply transformation
gg_bottomleft = proj_transformer_back.transform(
lim_lng_min_crop, lim_lat_min_crop)
gg_topright = proj_transformer_back.transform(
lim_lng_max_crop, lim_lat_max_crop)
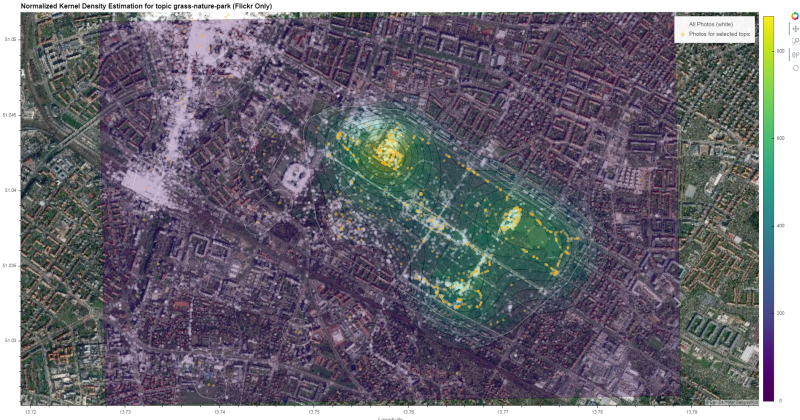
Construct final overlay from individual layers
- Backgorund Tiles (sattelite)
- All Points (Photos)
- KDE Contours
- Selected topic points ('green' photos)
- HDBSCAN Cluster Shapes
bg_layer = gv.tile_sources.EsriImagery.opts(alpha=1.0)
all_photos_layer = gv.Points(
(xy_list[1][0], xy_list[1][1]),
label='All Photos (white)',
crs=cc_proj).opts(size=5, color='white', alpha=0.3)
contours_layer = contours.opts(
colorbar=True, cmap='viridis', alpha=0.3,
clipping_colors={'NaN': (0, 0, 0, 0)})
topic_layer = gv.Points(
(xy_list[0][0], xy_list[0][1]),
label='Photos for selected topic',
crs=cc_proj).opts(size=5, color='orange', alpha=0.5)
cluster_layer = gv_shapes.opts(
line_color='white', line_width=0.5, fill_alpha=0)
Compile final output
Have a look at the generated interactive html here
gv_layers = hv.Overlay(
bg_layer * \
all_photos_layer * \
contours_layer * \
topic_layer * \
cluster_layer).opts(
responsive=True,
title=f"Frequentation surface for topic {topic}",
hooks=[set_active_tool],
data_aspect=0.6, # either set aspect ratio
# height=700, # or a fixed height
xlim=(gg_bottomleft[0], gg_topright[0]),
ylim=(gg_bottomleft[1], gg_topright[1])
)
gv_layers
Save to external HTML.
hv.save(
gv_layers,
OUTPUT / f'heatmap_freq_{topic}.html', backend='bokeh')
Optionally Modify data_aspect
- Because we're using explicit x-y-lim parameter, we need to specify the
data_aspectparameter, to stretch the output map to max width and a certain height - Depending on you data-area-rectangle, you may need to modify the parameter so that the height of the output map does not stretch beyond typical screen sizes
- Smaller
data_aspectvalues will result in a reduced height of the map - Alternatively, you can also remove
data_aspectand use themin_heightparameter (e.g.:min_height=600)
import flopy
from flopy.export.utils import export_contourf
epsg_output = tm.clusterer[TOPICS].crs_proj.split(":")[1]
export_contourf(
str(OUTPUT / '2021-07-12_KDE.shp'),
CF,
epsg=epsg_output)
Alpha Shapes
from typing import List, Dict, Any
def write_shapefile(
name: Path, schema: Dict[str, str],
properties_dict, geometries: List[Any],
epsg_code: str):
"""Write list of geometries to shapefile"""
with fiona.open(
name, 'w', 'ESRI Shapefile', schema,
crs=epsg_code) as shapefile:
for geometry in geometries:
shapefile.write({
'geometry': mapping(geometry),
'properties': properties_dict,
})
import fiona
from fiona.crs import from_epsg
from shapely.geometry import mapping, Polygon, Point
from datetime import date
today = str(date.today())
# Define a polygon feature geometry with one attribute
schema = {
'geometry': 'Polygon',
'properties': {'id': 'str'},
}
# Write a new Shapefile
write_shapefile(
name=OUTPUT / f'{today}_Clusters.shp',
schema=schema,
properties_dict={'id': "photo cluster"},
geometries=shape_list,
epsg_code=from_epsg(epsg_output))
Post points
Use list comprehension to create a list of Shapely points
points = [Point(xx, yy) for xx, yy in zip(xy_list[1][0], xy_list[1][1])]
schema = {
'geometry': 'Point',
'properties': {'id': 'str'},
}
write_shapefile(
name=OUTPUT / f'{today}_AllPhotos.shp',
schema=schema,
properties_dict={'id': "All photos"},
geometries=points,
epsg_code=from_epsg(epsg_output))
Write topic points:
points = [Point(xx, yy) for xx, yy in zip(xy_list[0][0], xy_list[0][1])]
write_shapefile(
name=OUTPUT / f'{today}_SelectedPhotos.shp',
schema=schema,
properties_dict={'id': "Photos for selected topic"},
geometries=points,
epsg_code=from_epsg(epsg_output))
Create Notebook HTML¶
!jupyter nbconvert --to html \
--output-dir=./out/ ./03_tagmaps.ipynb \
--template=../nbconvert.tpl \
--ExtractOutputPreprocessor.enabled=False >&- 2>&-
Create ZIP with all results¶
During the workshop, you have created several files in the out folder.
Files can be downloaded individually, but we can also zip contents, to make life easier.
%%time
zip_file = OUTPUT / f'{today}_workshop_results.zip'
if not zip_file.exists():
tools.zip_dir(OUTPUT, zip_file)
Download the zipfile from the out folder on the left.
Clean up input folder
tools.clean_folders([INPUT, Path.cwd() / "02_Output"])
Summary¶
Notes
- Social Media data is noisy
- Through filtering and aggregation, summary visualizations can be generated
- The Heat Map produced here is one example
- In the second part of the workshop, at the end of January, we'll visualize the clustered tagmaps data using ArcMap
Further work
- You have seen a number of data processing and visualization techniques in this workshop
- Before jumping on to other techniques, try to get familiar with package management and other tools for python development. Some suggestions are:
- Using Windows Subsystem for Linux (WSL) (if you're working in Windows)
- Using (Mini)Conda and Git in WSL
- Try our Jupyter Lab Docker Container (in WSL!)
- This was an experimental workshop. Tell us what you think could be improved (or what you liked), and if we should repeat something similar in the following semesters.
Please shutdown your notebook server
- if you're finished working through the notebooks
- Please shut down your Jupyter Hub server
- File > Shut Down
root_packages = [
'python', 'geoviews', 'holoviews', 'ipywidgets', 'descartes', 'hvplot',
'flopy', 'mapclassify', 'memory_profiler', 'python-dotenv', 'shapely',
'tagmaps', 'matplotlib', 'sklearn', 'numpy', 'pandas', 'bokeh', 'fiona']
tools.package_report(root_packages)